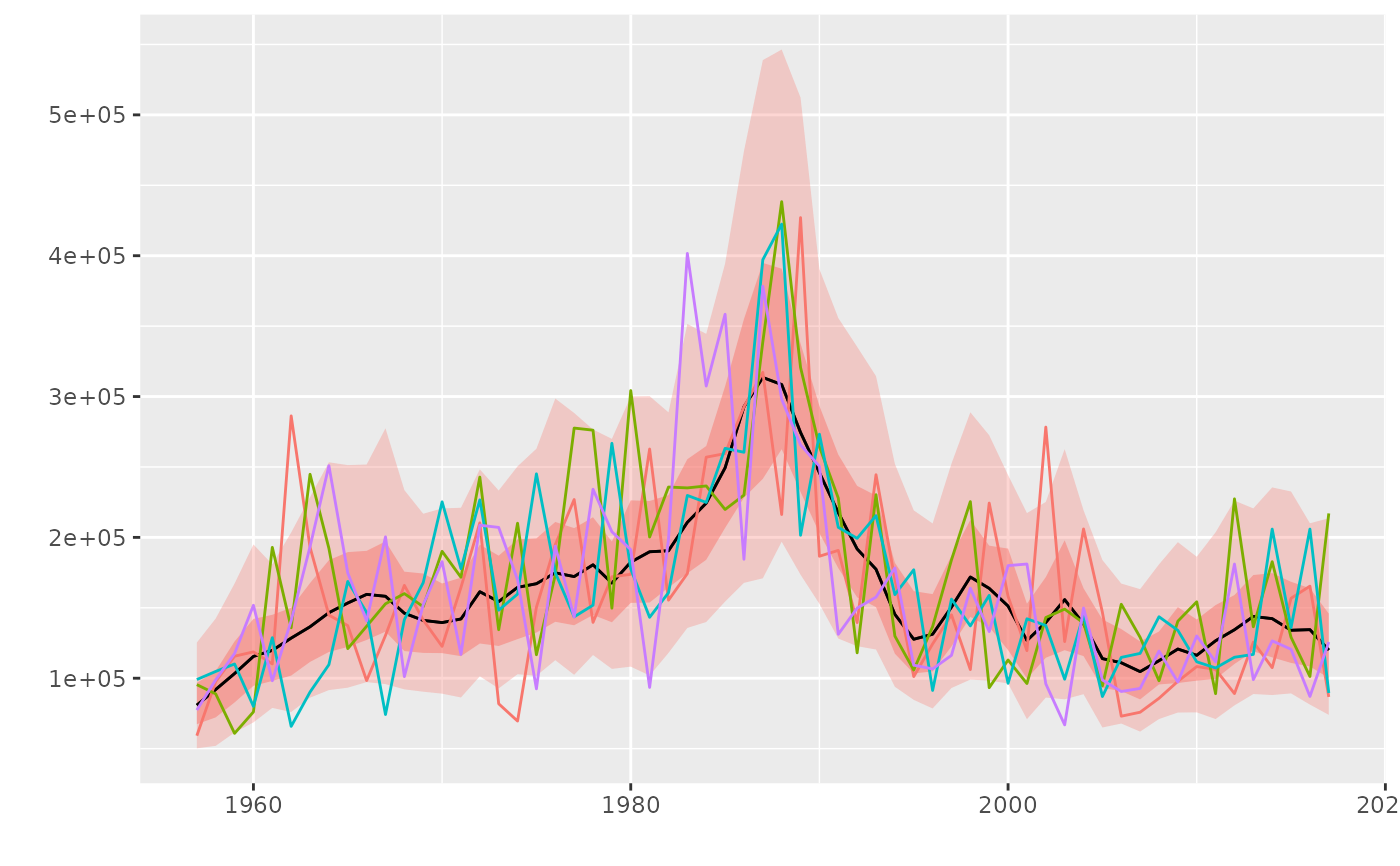
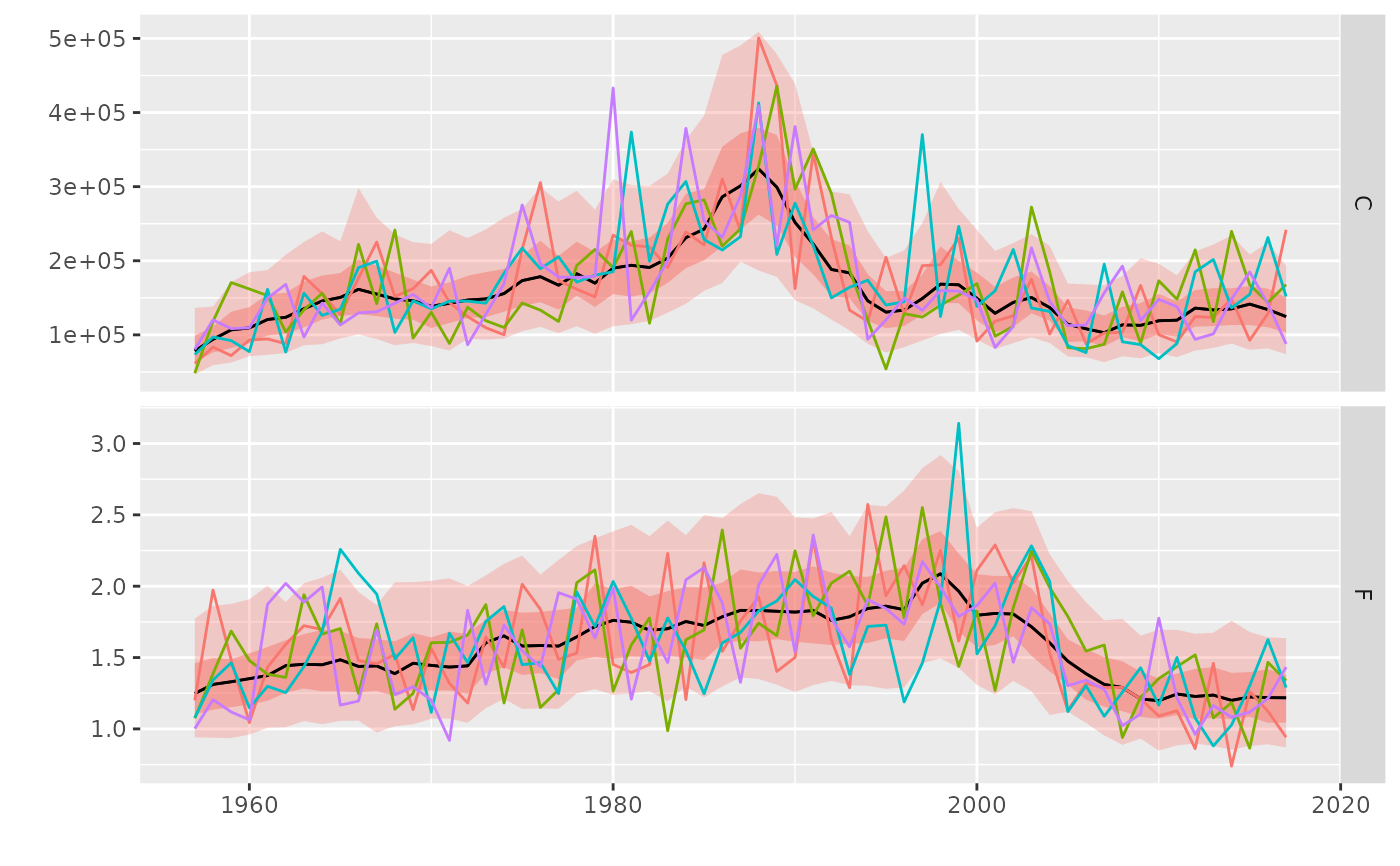
A geom for adding worms to probability intervals from geom_flquantiles
Source:R/geom.R
geom_worm.RdA geom for adding worms to probability intervals from geom_flquantiles
geom_worm(
data,
mapping = aes(colour = iter),
...,
stat = "identity",
position = "identity",
na.rm = FALSE
)Arguments
- data
Subset of data, select from full object using iter().
- mapping
Set of aesthetic mappings created by
aes(). If specified andinherit.aes = TRUE(the default), it is combined with the default mapping at the top level of the plot. You must supplymappingif there is no plot mapping.- ...
Other arguments passed on to
layer(). These are often aesthetics, used to set an aesthetic to a fixed value, likecolour = "red"orsize = 3. They may also be parameters to the paired geom/stat.- stat
The statistical transformation to use on the data for this layer, either as a
ggprotoGeomsubclass or as a string naming the stat stripped of thestat_prefix (e.g."count"rather than"stat_count")- position
Position adjustment, either as a string naming the adjustment (e.g.
"jitter"to useposition_jitter), or the result of a call to a position adjustment function. Use the latter if you need to change the settings of the adjustment.- na.rm
If
FALSE, the default, missing values are removed with a warning. IfTRUE, missing values are silently removed.
Aesthetics
`geom_worm` understands the following aesthetics (required aesthetics are in bold): - `colour` - `linetype` - `linewidth`