Fits Stock Recruitment Relationships (SRR) in TBM
srrTMB.RdFits Stock Recruitment Relationships (SRR) in TBM
srrTMB(
object,
spr0 = "missing",
s = NULL,
s.est = TRUE,
s.logitsd = 20,
r0.pr = "missing",
lplim = 0.001,
uplim = 0.3,
plim = lplim,
pmax = uplim,
nyears = NULL,
report.sR0 = FALSE,
inits = NULL,
lower = NULL,
upper = NULL,
SDreport = TRUE,
verbose = FALSE
)Arguments
- object
Input FLSR = as.FLSR(stock,model) object with current model options
bevholtSV
rickerSV
segreg
geomean
- spr0
unfished spawning biomass per recruit from FLCore::spr0(FLStock)
- s
steepness parameter of SRR (fixed or prior mean)
- s.est
option to estimate steepness
- s.logitsd
prior sd for logit(s), default is 1.4 (flat) if s.est = TRUE
- r0.pr
option to condition models on r0 priors (NULL = geomean)
- lplim
lower bound of spawning ratio potential SRP, default 0.0001
- uplim
upper bound of plausible spawning ratio potential SRP , default 0.3
- plim
depreciated plim = usrp
- pmax
depreciated pmax = lsrp
- nyears
yearMeans from the tail used to compute a,b from the reference spr0 (default all years)
- report.sR0
option to report s and R0 instead of a,b
- inits
option to specify initial values of log(r0), log(SigR) and logit(s)
- lower
option to specify lower bounds of log(r0), log(SigR) and logit(s)
- upper
option to specify upper bounds of log(r0), log(SigR) and logit(s)
- SDreport
option to converge hessian and get vcov
- verbose
if TRUE, it shows tracing
Value
A list containing elements 'FLSR', of class FLSR
Examples
data(ple4)
gm <- srrTMB(as.FLSR(ple4,model=geomean),spr0=mean(spr0y(ple4)))
bh <- srrTMB(as.FLSR(ple4,model=bevholtSV),spr0=spr0y(ple4))
ri <- srrTMB(as.FLSR(ple4,model=rickerSV),spr0=spr0y(ple4))
hs <- srrTMB(as.FLSR(ple4,model=segreg),spr0=spr0y(ple4),lplim=0.05,uplim=0.2)
srs = FLSRs(gm=gm,bh=bh,ri=ri,hs=hs) # combine
plotsrs(srs)
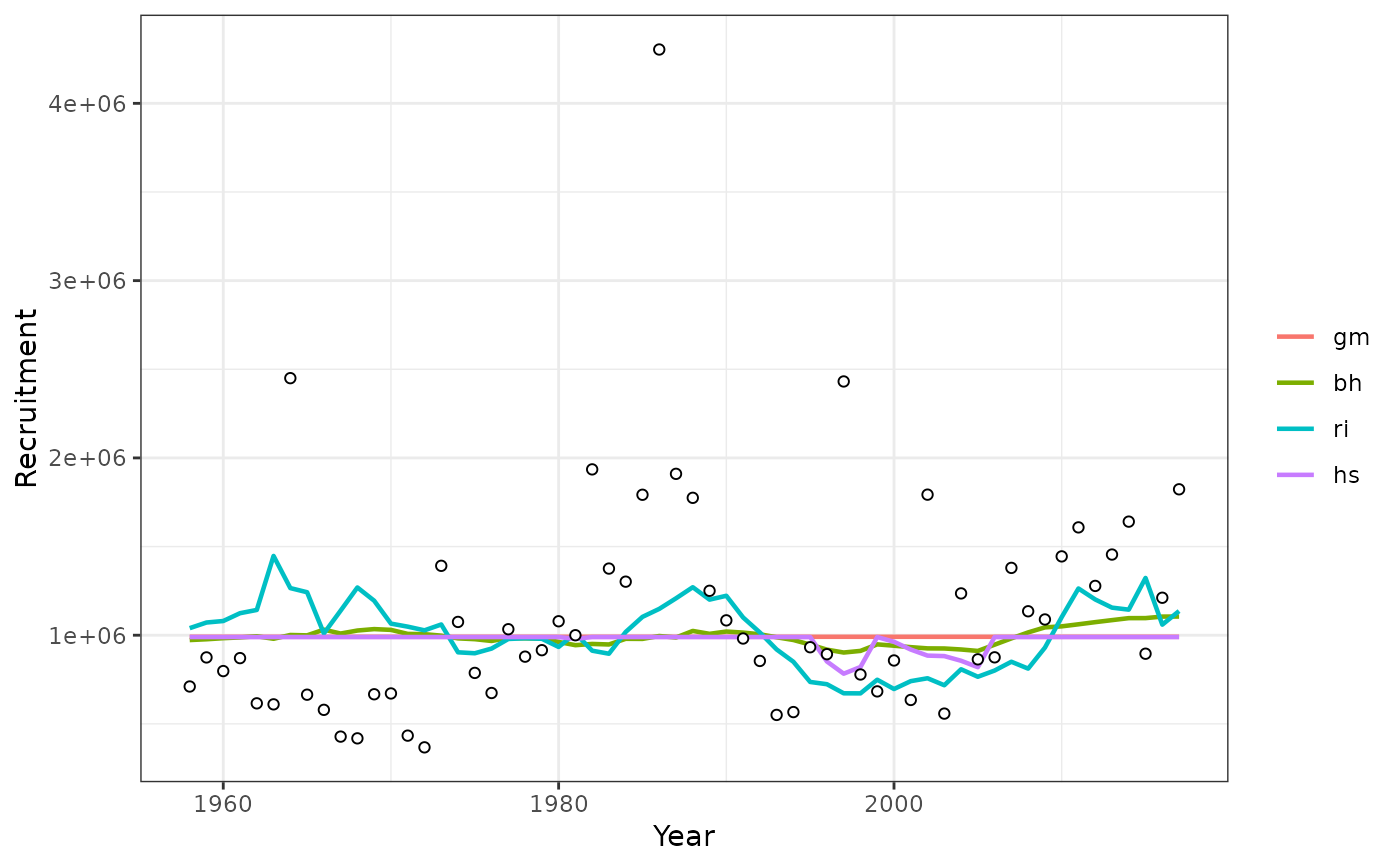
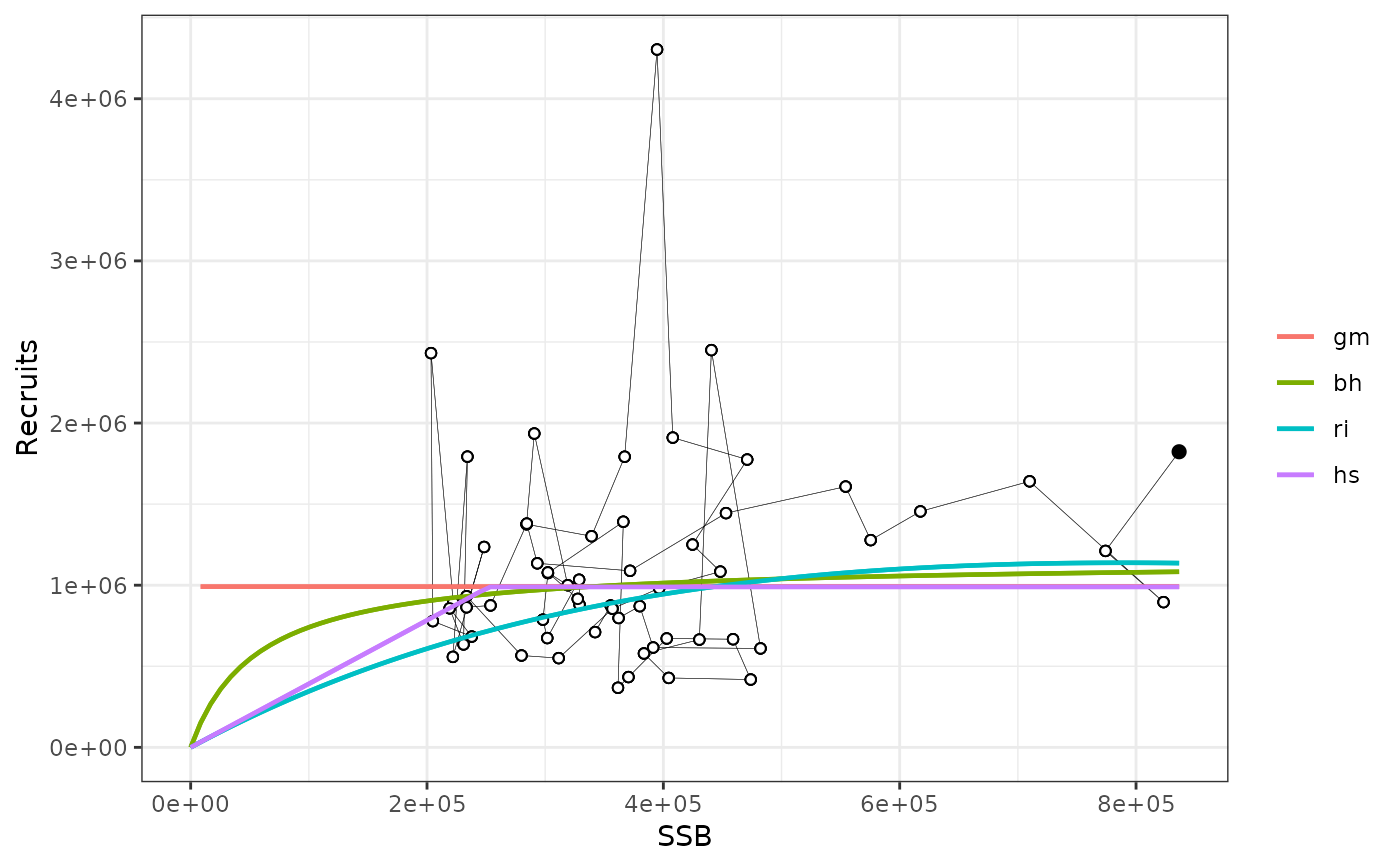 plotsrts(srs) # relative
plotsrts(srs) # relative
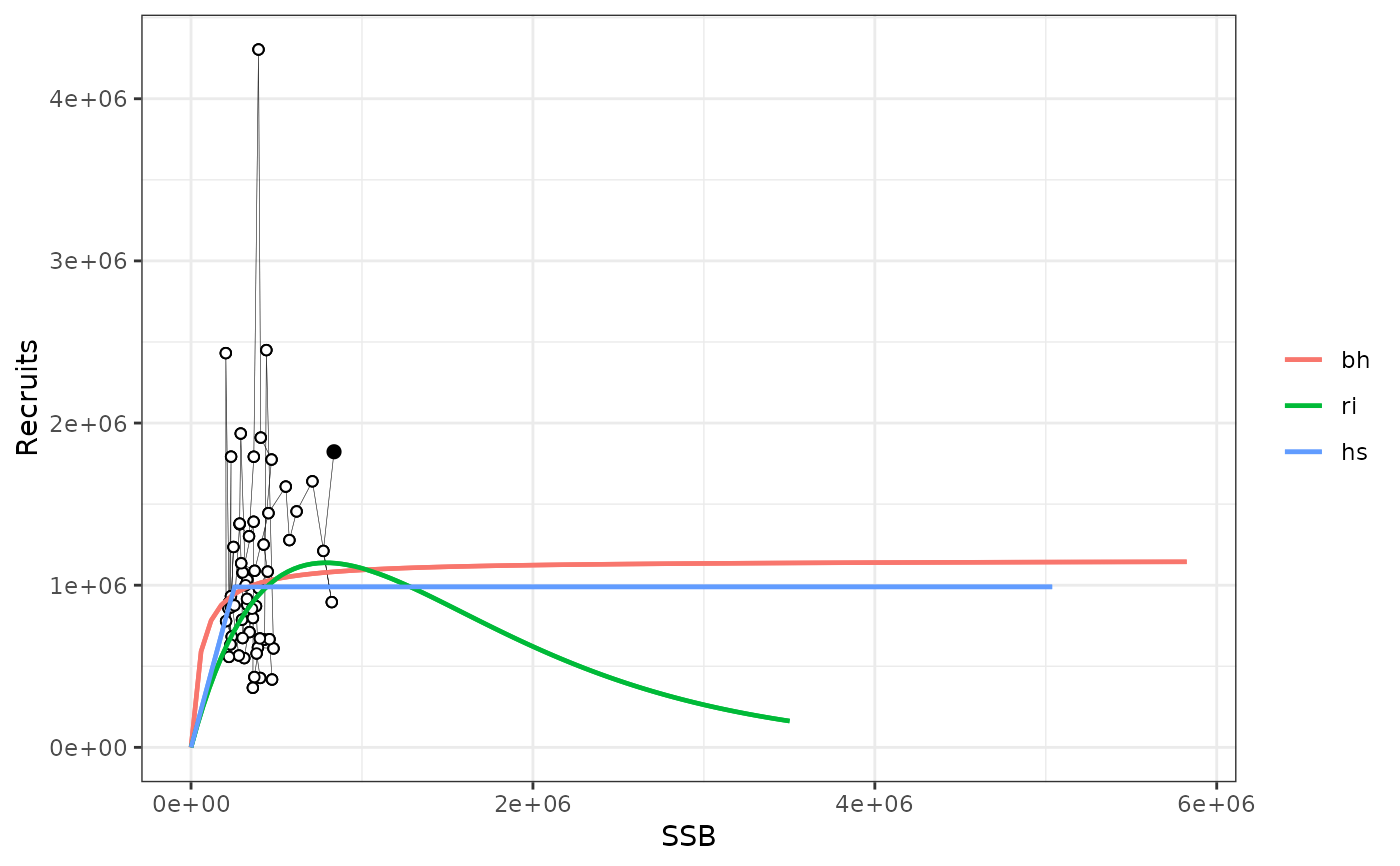
 plotsrs(srs[2:4],b0=TRUE) # through to B0
plotsrs(srs[2:4],b0=TRUE) # through to B0
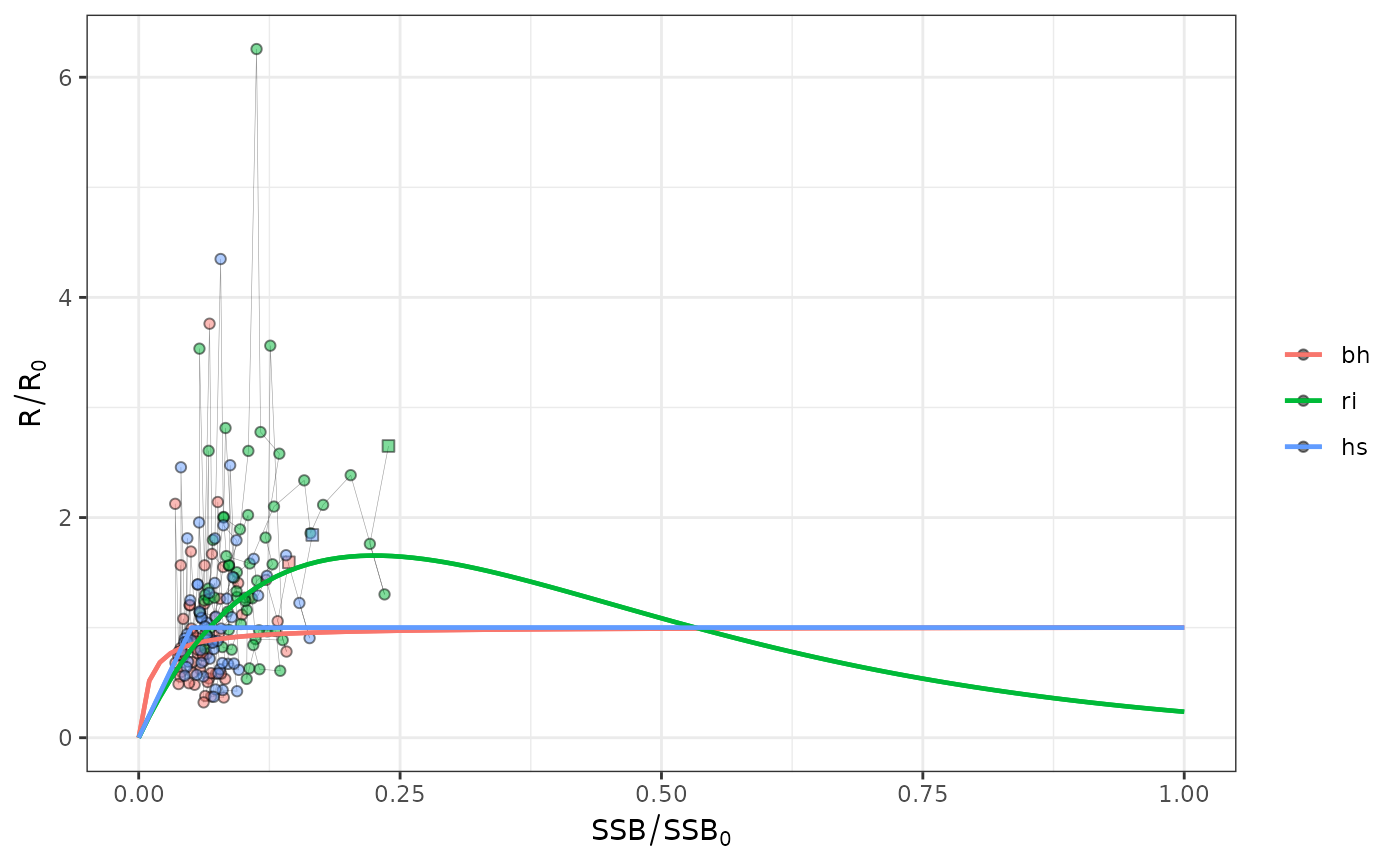
 plotsrs(srs[2:4],b0=TRUE,rel=TRUE) # relative
plotsrs(srs[2:4],b0=TRUE,rel=TRUE) # relative
 gm@SV # estimates
#> s sigmaR R0 rho B0
#> 1 NA 0.4827602 991110.1 0.4088983 5020664
do.call(rbind,lapply(srs,AIC))
#> [,1]
#> gm 85.87597
#> bh 87.37908
#> ri 93.09520
#> hs 89.81315
gm@SV # estimates
#> s sigmaR R0 rho B0
#> 1 NA 0.4827602 991110.1 0.4088983 5020664
do.call(rbind,lapply(srs,AIC))
#> [,1]
#> gm 85.87597
#> bh 87.37908
#> ri 93.09520
#> hs 89.81315